Code
demo_r_mlifexp %>%
left_join(unit, by = "unit") %>%
group_by(unit, Unit) %>%
summarise(Nobs = n()) %>%
arrange(-Nobs) %>%
{if (is_html_output()) print_table(.) else .}| unit | Unit | Nobs |
|---|---|---|
| YR | Year | 3351131 |
Data - Eurostat
demo_r_mlifexp %>%
left_join(unit, by = "unit") %>%
group_by(unit, Unit) %>%
summarise(Nobs = n()) %>%
arrange(-Nobs) %>%
{if (is_html_output()) print_table(.) else .}| unit | Unit | Nobs |
|---|---|---|
| YR | Year | 3351131 |
demo_r_mlifexp %>%
left_join(sex, by = "sex") %>%
group_by(sex, Sex) %>%
summarise(Nobs = n()) %>%
arrange(-Nobs) %>%
{if (is_html_output()) print_table(.) else .}| sex | Sex | Nobs |
|---|---|---|
| T | Total | 1117101 |
| F | Females | 1117015 |
| M | Males | 1117015 |
demo_r_mlifexp %>%
left_join(age, by = "age") %>%
group_by(age, Age) %>%
summarise(Nobs = n()) %>%
arrange(-Nobs) %>%
{if (is_html_output()) datatable(., filter = 'top', rownames = F) else .}demo_r_mlifexp %>%
left_join(geo, by = "geo") %>%
group_by(geo, Geo) %>%
summarise(Nobs = n()) %>%
arrange(-Nobs) %>%
{if (is_html_output()) datatable(., filter = 'top', rownames = F) else .}demo_r_mlifexp %>%
filter(time == "2018",
nchar(geo) == 4,
sex == "T",
age == "Y1") %>%
left_join(geo, by = "geo") %>%
select(geo, Geo, values) %>%
right_join(europe_NUTS2, by = "geo") %>%
filter(long >= -15, lat >= 33) %>%
ggplot(., aes(x = long, y = lat, group = group, fill = values)) +
geom_polygon() + coord_map() +
scale_fill_viridis_c(na.value = "white",
direction = -1,
breaks = seq(70, 90, 2),
values = c(0, 0.1, 0.3, 0.5, 0.7, 0.9, 1)) +
theme_void() + theme(legend.position = c(0.25, 0.85)) +
labs(fill = "Life Expectancy (Men and Women)")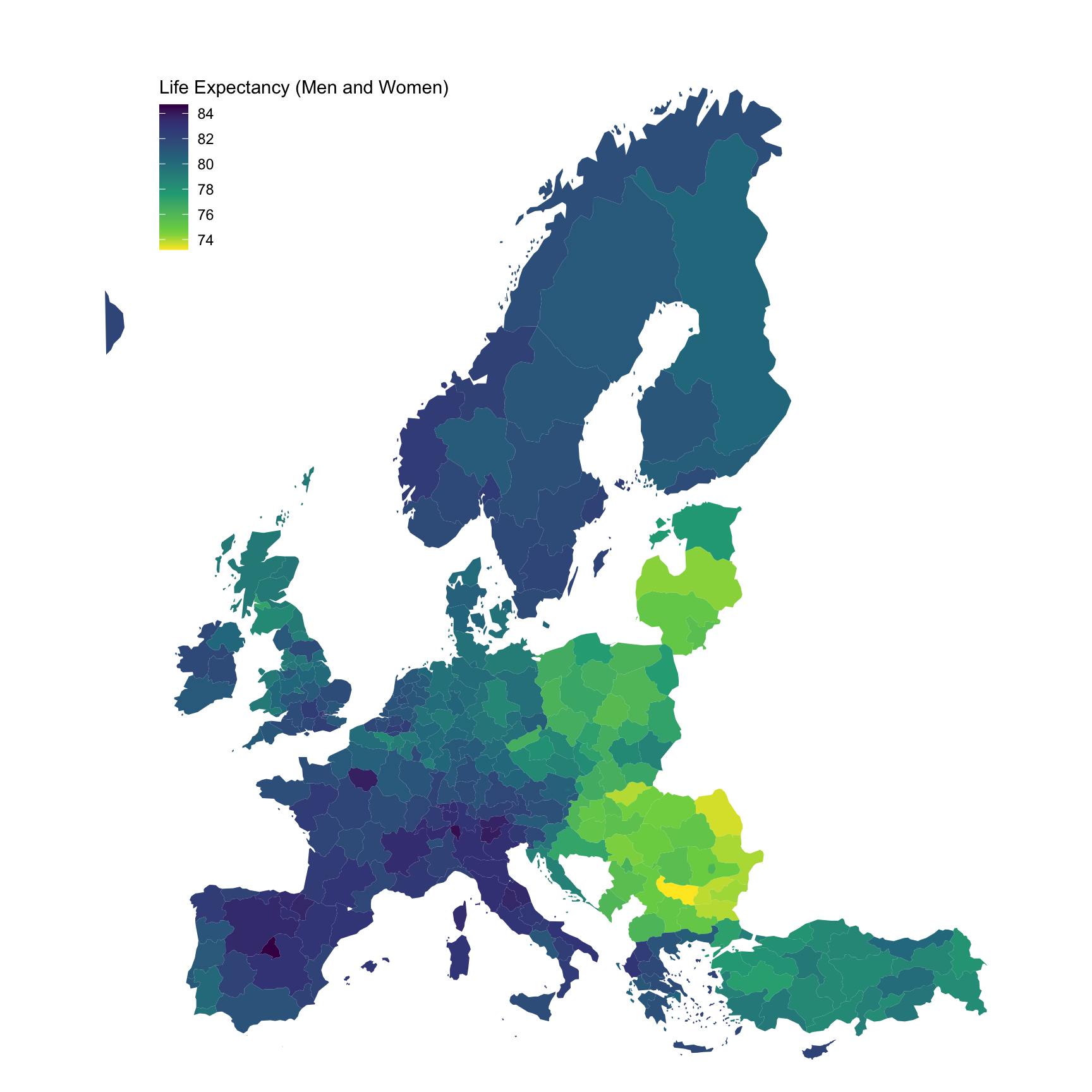
demo_r_mlifexp %>%
filter(time == "2018",
nchar(geo) == 4,
sex == "M",
age == "Y1") %>%
left_join(geo, by = "geo") %>%
select(geo, Geo, values) %>%
right_join(europe_NUTS2, by = "geo") %>%
filter(long >= -15, lat >= 33) %>%
ggplot(., aes(x = long, y = lat, group = group, fill = values)) +
geom_polygon() + coord_map() +
scale_fill_viridis_c(na.value = "white",
direction = -1,
breaks = seq(70, 90, 2),
values = c(0, 0.1, 0.3, 0.5, 0.7, 0.9, 1)) +
theme_void() + theme(legend.position = c(0.25, 0.85)) +
labs(fill = "Life Expectancy (Men)")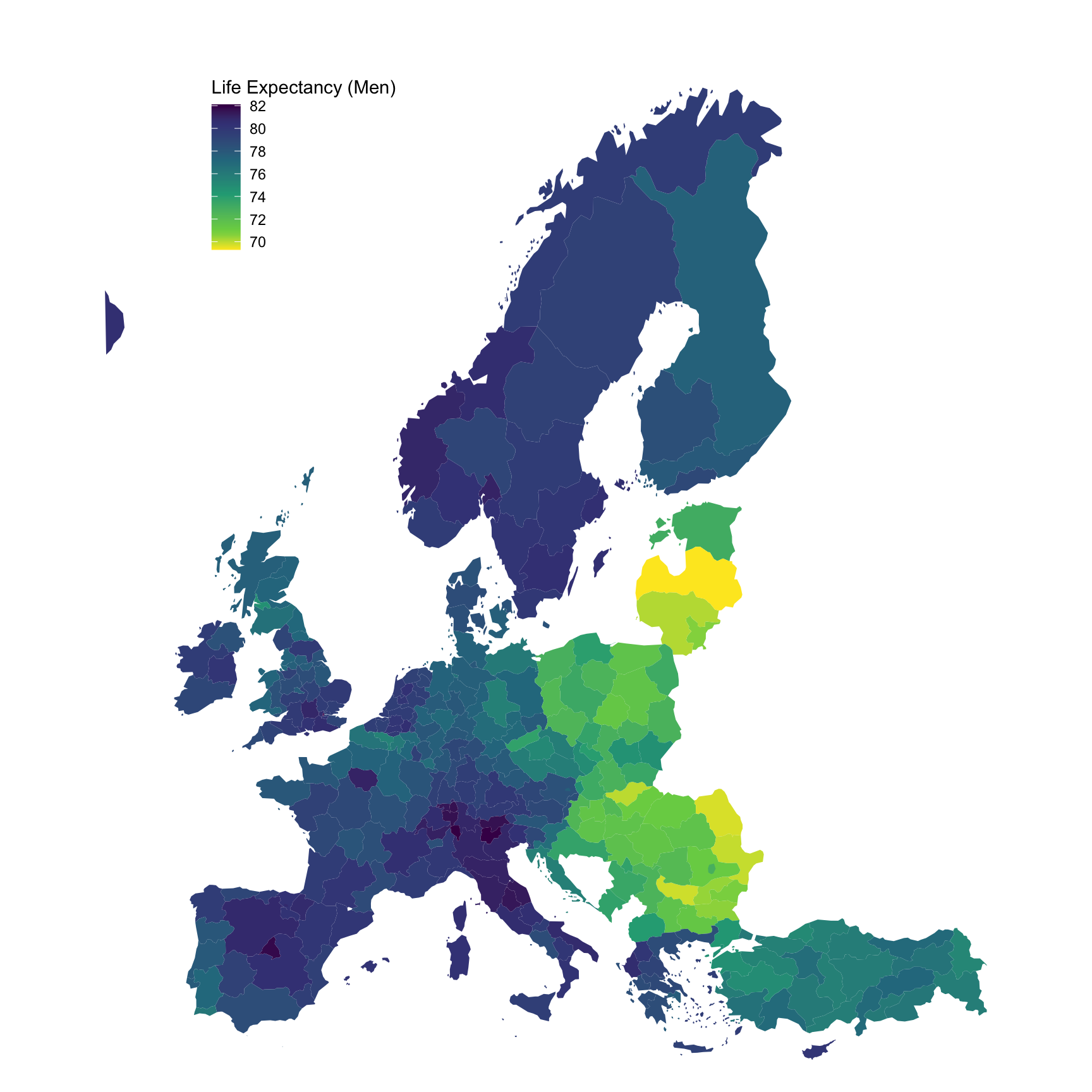
demo_r_mlifexp %>%
filter(time == "2018",
nchar(geo) == 4,
sex == "F",
age == "Y1") %>%
left_join(geo, by = "geo") %>%
select(geo, Geo, values) %>%
right_join(europe_NUTS2, by = "geo") %>%
filter(long >= -15, lat >= 33) %>%
ggplot(., aes(x = long, y = lat, group = group, fill = values)) +
geom_polygon() + coord_map() +
scale_fill_viridis_c(na.value = "white",
direction = -1,
breaks = seq(70, 90, 2),
values = c(0, 0.1, 0.3, 0.5, 0.7, 0.9, 1)) +
theme_void() + theme(legend.position = c(0.25, 0.85)) +
labs(fill = "Life Expectancy (Women)")